The Simulation Experiment Description Markup Language (SED-ML) is a format for the standardized encoding of simulation setups of computational models in biology (e.g., in SBML or CellML format). SED-ML-encoded experiments are reproducible. The project is a community effort under the umbrella of the COMBINE initiative.
We argue that in order to reuse models from public databases, such as BioModels Database or the Physiome Model Repository, these models should be provided together with applicable simulation setups. Therefore, SED-ML DB is a project to develop an integrated storage approach for models, simulation setups and simulation results. The aim of this subproject is to make SED-ML files searchable, provide version control, and to directly link SED-ML outputs to model entities in the model database.
SED-ML is an XML format, developed as a community effort. Since XML has a graph-like structure, it is advantageous to use a graph database as storage model. We use the open-source, high-performance, enterprise-grade NOSQL graph database Neo4j. Neo4j is a robust (fully ACID) transactional property graph database. Due to its graph data model, Neo4j is agil and fast.
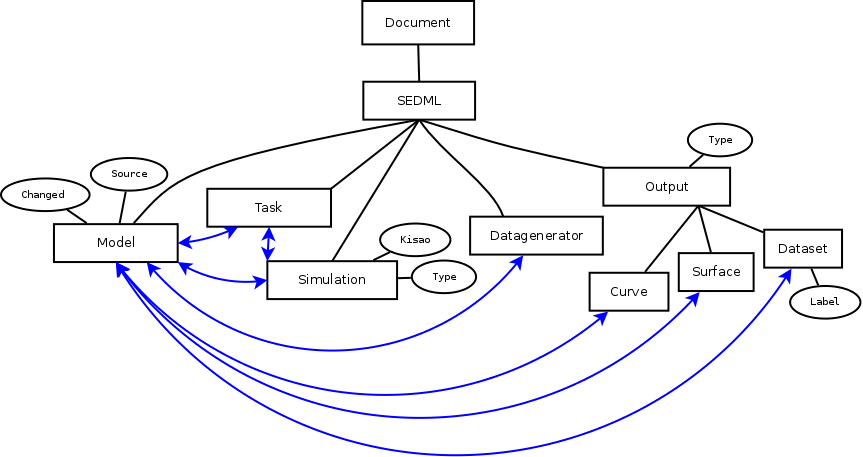
SED-ML schema for Neo4j implementation: Entities in boxes characterize graph nodes, ellipses identify attributes, lines indicate relationships. Black lines mark the “belongs_to” relationship, blue lines identify more complex relationships.
Brought to you by