Read more about BiVeS in our preprint An algorithm to detect and communicate the differences in computational models describing biological systems and consult our tracs for more details on BiVeS.
The Input
As an input BiVeS expects two versions of a model encoded in standard formats. We currently support models encoded in either SBML or CellML.
The Algorithm
The algorithm implemented in BiVeS maps the entities of the model documents onto each other and identifies inserted, deleted, moved, and updated element.
The Output
BiVeS is able to export identified differences in both machine- and human-readable formats.
- The XML encoded delta contains all differences for a subsequent processing by machines
- The highlighted reaction network displays the differences graphically, which makes is easy to grasp the changes. The graph is encoded in GraphML, DOT, or JSON.
- The text-based report lists all differences between both versions. It is encoded in either MarkDown, reStructuredText, or HTML.
Usage
BiVeS is intended to be used as a library in other software projects. It provides an Java API (more information on the API) to compare models directly from within your Java projects. Tools not developed in Java may use the BiVeS web service (more information on the web service and a client library) or call BiVeS directly from command line.
Download
The most current binaries of BiVeS can be downloaded from our bin-repository at bin.sems.uni-rostock.de.
Dev
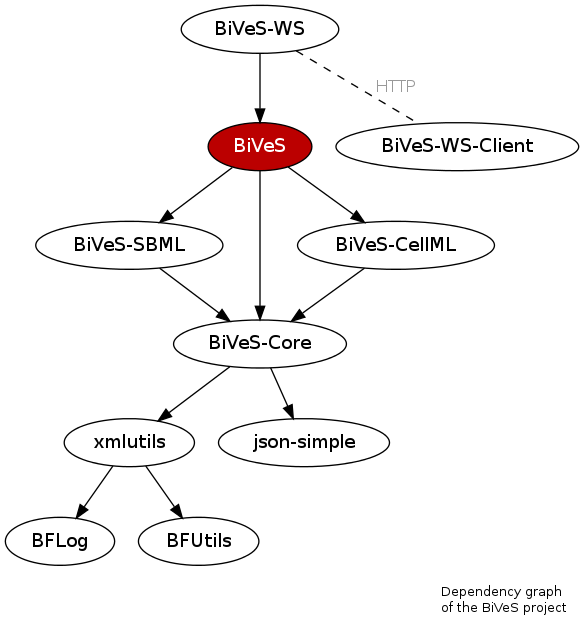
BiVeS is developed in Java. It consists of different modules, as shown in Figure 1. Each module has its own git repository and a trac to manage the development:
- BiVeS-Core:
git clone https://github.com/SemsProject/BiVeS-Core.git - BiVeS-SBML:
git clone https://github.com/SemsProject/BiVeS-SBML.git - BiVeS-CellML:
git clone https://github.com/SemsProject/BiVeS-CellML.git - BiVeS-FrameWork:
git clone https://github.com/SemsProject/BiVeS.git - BiVeS-WebApp:
git clone https://github.com/SemsProject/BiVeS-WS.git - BiVeS-WebApp Client:
git clone https://github.com/SemsProject/BiVeS-WS-Client.git
Besides the sources you’ll find an ant build file and a maven pom.xml.
To build the project using ant create a directory lib and copy the binaries of the dependencies to it. Then, just call ant dist to build the library.
To build the project using maven just call mvn package to let maven download dependencies and compile the project. Don’t forget to include our maven repository to your list of repositories.
Our trac contains further information on how to build and use BiVeS. And logos in different formats are available from our gallery.
Brought to you by