Vasundra Touré et al. published a paper on the STON framework in BMC Bioinformatics!
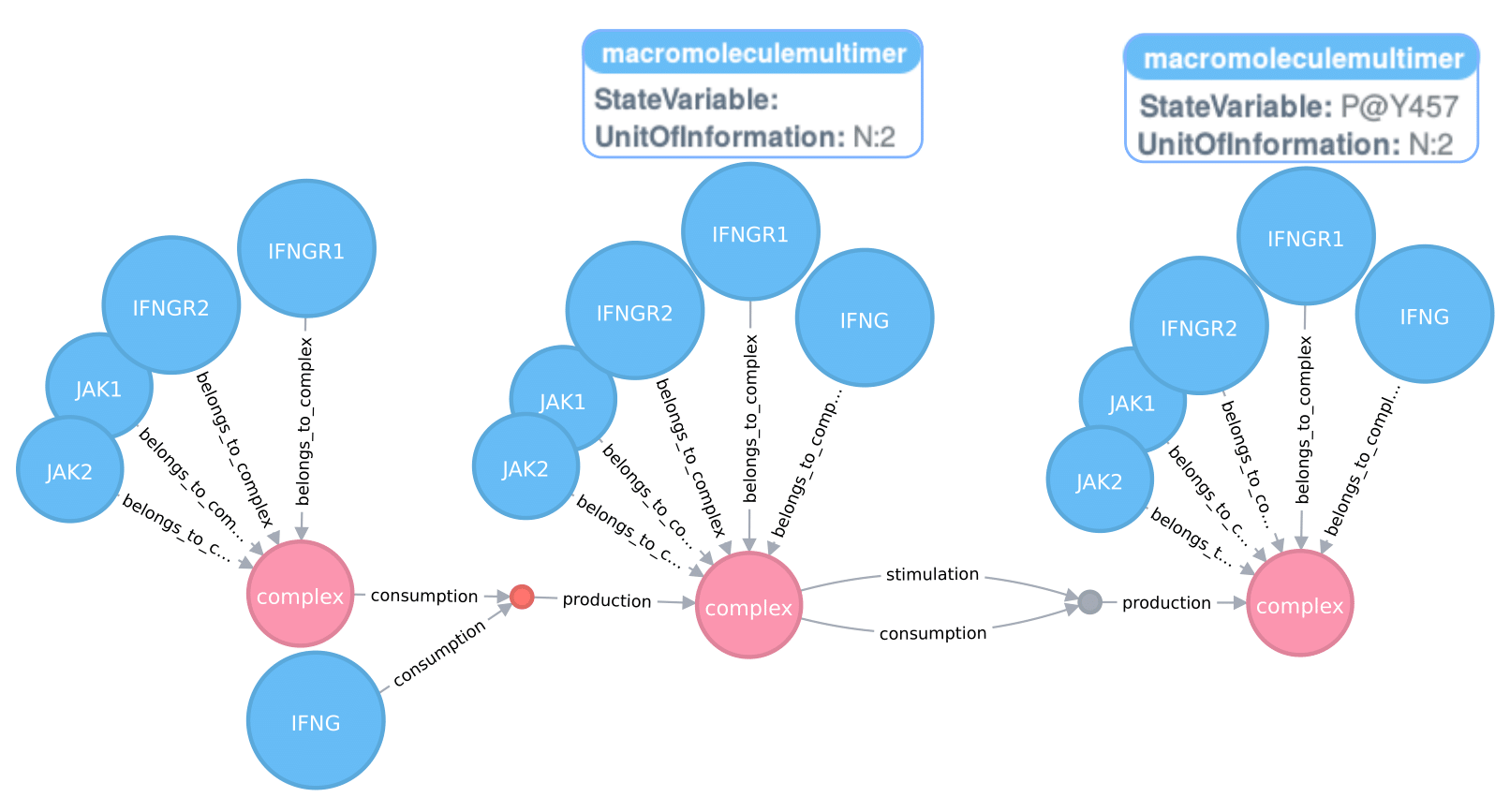
STON is meant to explore biological networks in SBGN using a graph database called Neo4j. The framework enables the query of SBGN maps.
Paper abstract:
Background
When modeling in Systems Biology and Systems Medicine, the data is often extensive, complex and heterogeneous. Graphs are a natural way of representing biological networks. Graph databases enable efficient storage and processing of the encoded biological relationships. They furthermore support queries on the structure of biological networks.
Results
We present the Java-based framework STON (SBGN TO Neo4j). STON imports and translates metabolic, signalling and gene regulatory pathways represented in the Systems Biology Graphical Notation into a graph-oriented format compatible with the Neo4j graph database.
Conclusion
STON exploits the power of graph databases to store and query complex biological pathways. This advances the possibility of: i) identifying subnetworks in a given pathway; ii) linking networks across different levels of granularity to address difficulties related to incomplete knowledge representation at single level; and iii) identifying common patterns between pathways in the database.
Read more at: http://www.biomedcentral.com/1471-2105/17/494.
The framework resources are available in sourceforge.